Below is a statement delivered November 2018 to the Euro-Bioimaging Industry
Board regarding the support of proprietary file formats by Bio-Formats. This
was discussed during the
From Images to Knowledge with ImageJ & Friends meeting in
December and since then, there have been a growing number of conversations
about a common format for bioimaging data. We're posting it here to tie the
conversations back together and continue an open discussion of this critical
issue.
As many of you know, work on Bio-Formats began in 2006, and over the first 10 years of development, support was added for over 140 file formats. If you include the per-format variants that have emerged over the years, that might be as much as 5 or 10 times higher, but precise numbers are difficult at best.
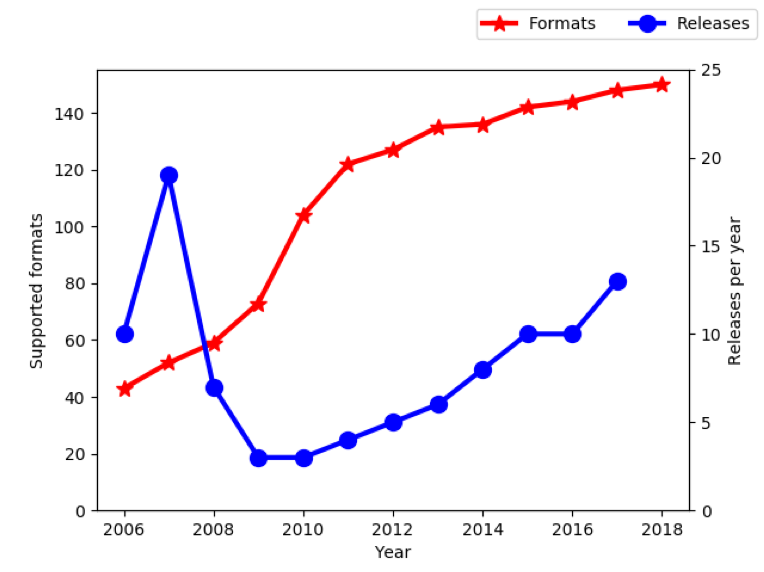
In 2016, we issued a public statement that OME, or more specifically its funding model, was not going to keep up with the accelerated development of new formats. We warned that we would be spending less time on closed formats, and we suggested that format developers either move to open formats or invest their own time or money to support their formats.
How did that turn out? Well, two years later the growth curve has naturally levelled off as we pursue other priorities. Currently there are just over 150 formats supported. One company, 3i, has taken over support of their own file format (Slidebook6) with a closed source reader that lives outside of Bio-Formats.
A few other companies have added support for their format either by contributing directly to the library or by commissioning Glencoe Software to do so. Where necessary, the open source team has added support for formats that are needed for their funded priorities like datasets published in the Image Data Resource.1,2,3,4
Paying for the initial cost of a format is not enough.
But paying for the initial cost of a format is not enough. The need for indefinite support carries a larger, longer-lived price tag that leaves data written in a given format constantly at risk. These costs are exacerbated by format variants. Even when a format is defined following standards like DICOM, there is a need to contend with multiple implementations as is the case in the radiology domain. The same happened with the Olympus OIR format added in 2017 in partnership with Olympus Europe. Following public release, the community has periodically reported breakages caused by new variants of the format. 5,6,7,8,9
Put simply, the format landscape has scaled beyond a manageable level.
Put simply, the format landscape has scaled beyond a manageable level. The result is that scientists end up blocked in accessing and properly handling their data, and thus blocked in their scientific endeavor. If Bio-Formats were to cease to exist, a large percentage of imaging data would immediately cease to be accessible at least until someone took on the burden of support.
We understand the push to develop new formats. From numerous interactions, we know how crucial it is for data producers to be able to write data quickly as well as it is for users to be able to access their data quickly, and both across as many platforms as possible. We also know that, optimally, this ecosystem should all just keep working for years to come. But while these requirements need to be fulfilled, something must give.
We think the only scalable way forward is to work together on an ever smaller number of formats.
We think the only scalable way forward is to work together on an ever smaller number of formats. That’s why we’ve been concentrating on open formats instead of adding new proprietary formats. For example, Bio-Formats 6.1 adds support for the open BigDataViewer (BDV) format, a strong candidate for support across the community.
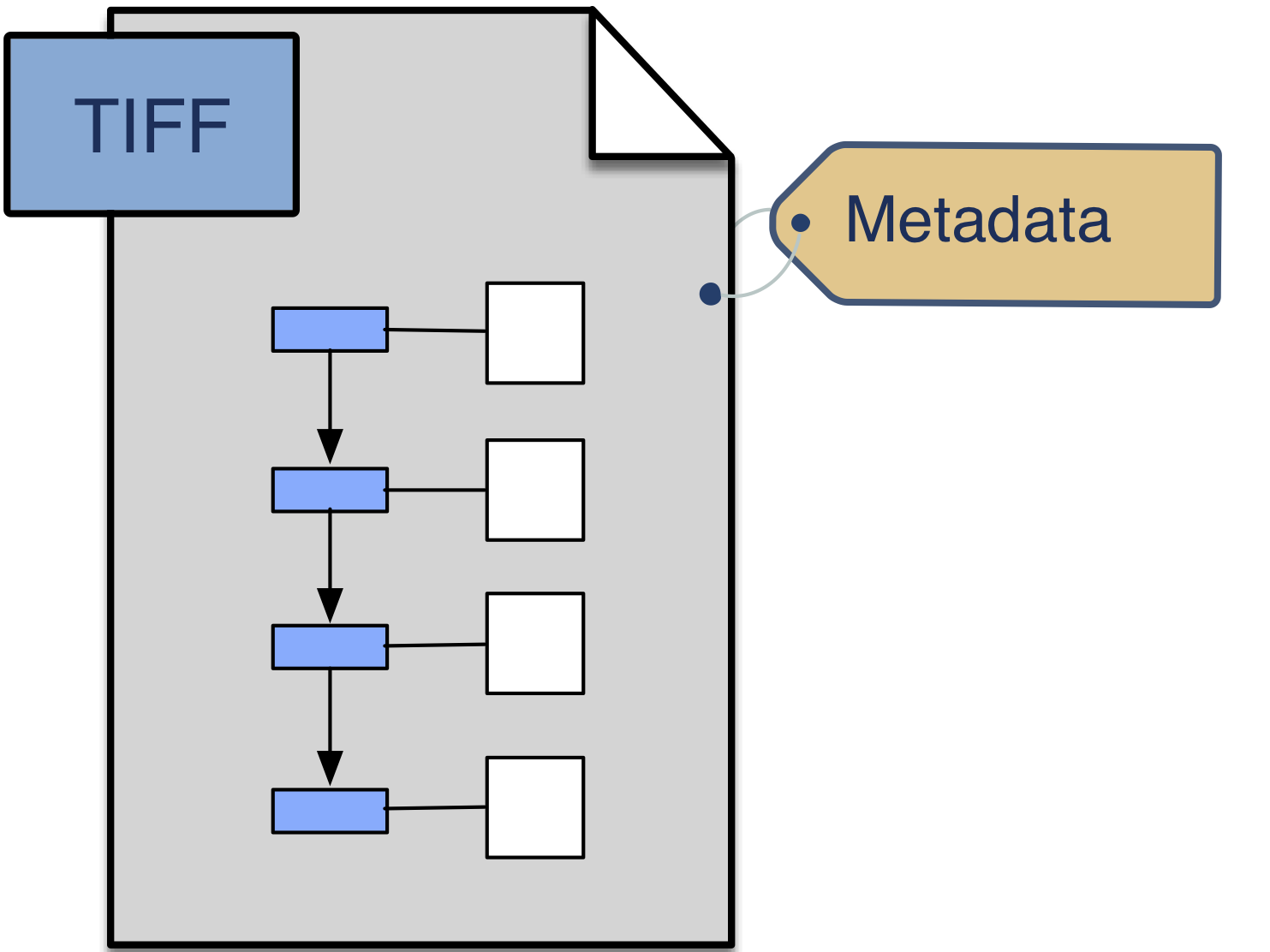
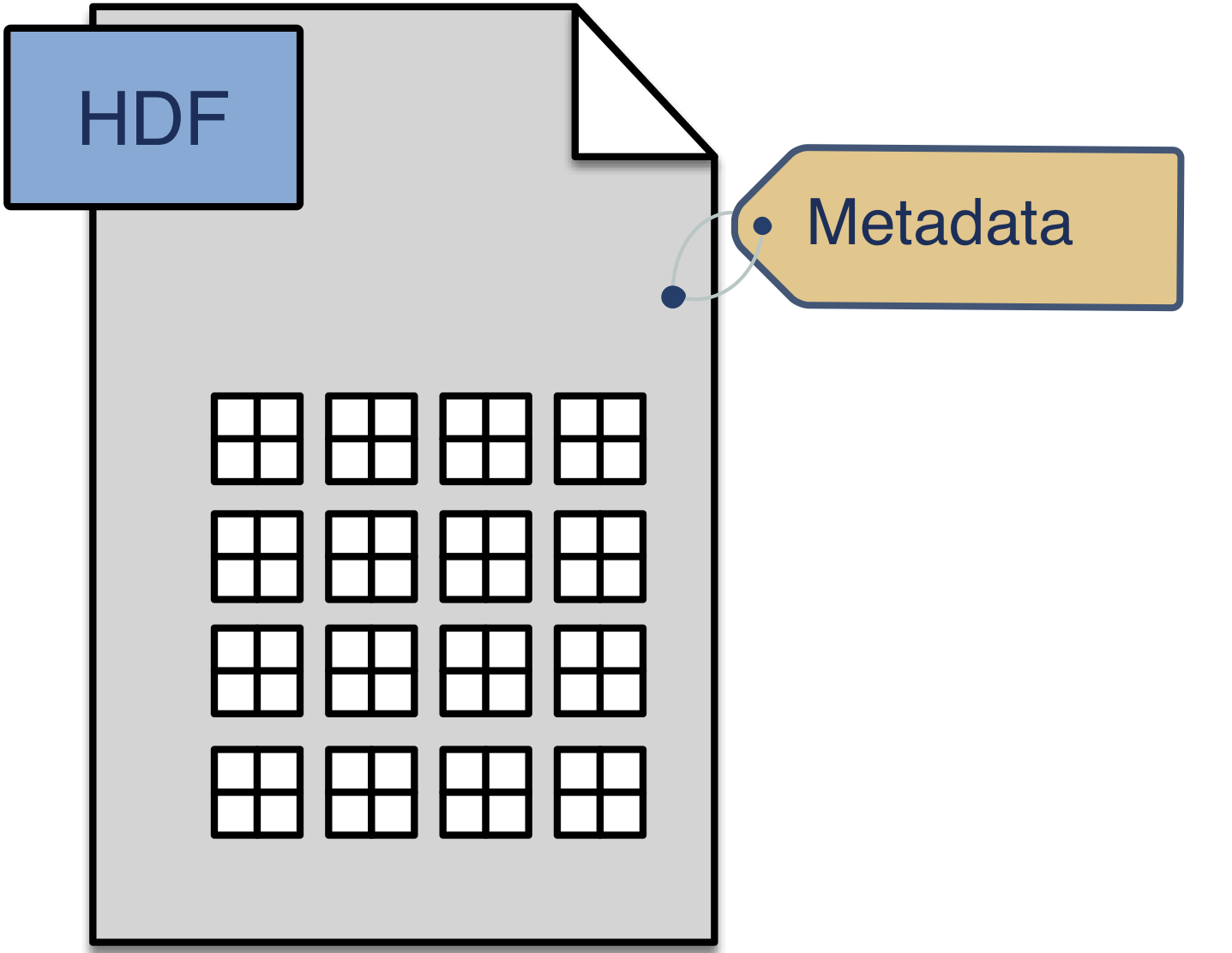
BDV provides a testbed for moving beyond the current single binary format of OME-TIFF. The OME Model will be extended to permit describing the multiscale, multidimensional data that is currently stored in BDV XML/H5. As a stable container format, HDF5 allows us a quick way to validate these concepts.
At the same time there’s a consensus that HDF5 itself as currently implemented cannot be the only binary container for our community, and, therefore, we are also collaborating on next-generation open-source, chunked (or “cloud”) formats for the scale of data generated by future acquisition systems. Two candidates — Zarr and N5 — were independently developed but overlap in most of their core concepts. Both communities have since begun work on a common storage spec, and other groups from NetCDF to Pangeo are getting involved.
We would like to see a community agreement between the various parties on a minimal set of open formats covering a broad range of imaging modalities.
We would like to see the bioimaging community agree on set of open formats covering a broad range of imaging modalities. We need to reduce long-term cost of our domain’s file formats and their variants. We want data users and producers to be able to ensure the long-term viability of their data.
OME-TIFF has been available for over a decade and today is in use by software across industry and academia, minimally as an export format, but it still doesn’t have the traction to stop a proliferation of new file formats. As support for this new binary format solidifies, we intend to invest long-term support in a new OME format.
Some of this work is the regular work of supporting the bioimaging community, but we feel this is a larger effort that could use more collaboration and funding. We are considering an application to the CZI’s Essential Open Source Software call and welcome any coordinated efforts. Beyond that, a truly common format will need indefinite support, and we will continue to look for avenues to do so.
You’re invited to discuss this forum post on the image.sc topic.
-
https://www.openmicroscopy.org/community/viewtopic.php?f=13&t=8360 ↩
-
https://www.openmicroscopy.org/community/viewtopic.php?f=13&t=8362 ↩
-
https://www.openmicroscopy.org/community/viewtopic.php?f=13&t=8522 ↩
-
https://list.nih.gov/cgi-bin/wa.exe?A2=IMAGEJ;c77987bc.1807 ↩
-
https://forum.image.sc/t/problems-opening-olympus-oir-files-using-bio-formats/24747 ↩

