Attending: Will, Jason, Jean-Marie, June, Mark, Melissa, Petr, Frances, Seb, Josh, David, Dom,
Kevin, Erin, Wilma, Chris, Emil
Start: 2:00 pm
-
Accepting minutes from last meeting
-
Project Status
(2-3 minutes each)
-
IDR:
-
Seb: release planned for tomorrow. 3 studies.
-
Idr0082 pathology, mouse (idr0092), HPA
-
But: Antibody annotations in incompatible state - fix tested
on pilot - looking good for release tomorrow.
-
-
-
NGFF:
-
Josh: few PRs opened recently
-
Can discuss after this meeting
-
Color handling proposals - see
omero-ms-zarr issue
-
-
OME-TIFF ‘slow’ (image.sc discussions)
-
-
OMERO 5
-
Josh: omero-py / omero-web releases upcoming?
-
Omero-py release when Simon is back
- Omero-web - fix for Qupath - need wider feedback on response
code change
- Seb: big changes coming. If this is a bug-fix (small
impact) then should get it out. Will can drive release.
- Seb: big changes coming. If this is a bug-fix (small
- Jason: good to get schedule defined
-
-
Mark: Deep copy PR is open
- Josh: tried to build whole OMERO stack in Docker using open
PRs -
- Much more work? Mark: not a whole lot. But lots of scenarios
to test
- Currently only testable via the CLI & API. May conflict with
web chown work.
- Concern over testing deep-copy on merge-ci - make snapshot
- Josh: tried to build whole OMERO stack in Docker using open
-
-
SA
-
Glencoe:
-
TileDB stuff being pushed to bioformats2raw
- Some upstream changes requested to improve portability
-
-
Community
-
AOB
(5 min. max; tech. Discussion should be highlighted to relevant people and rescheduled)
-
Scheduling of next week’s team meeting.
-
HPA talk at 2pm next Tuesday.
-
Go for 4pm - 4.30pm try to keep it short
-
Main Topic
(20-25 minutes plus 15 minutes questions max)
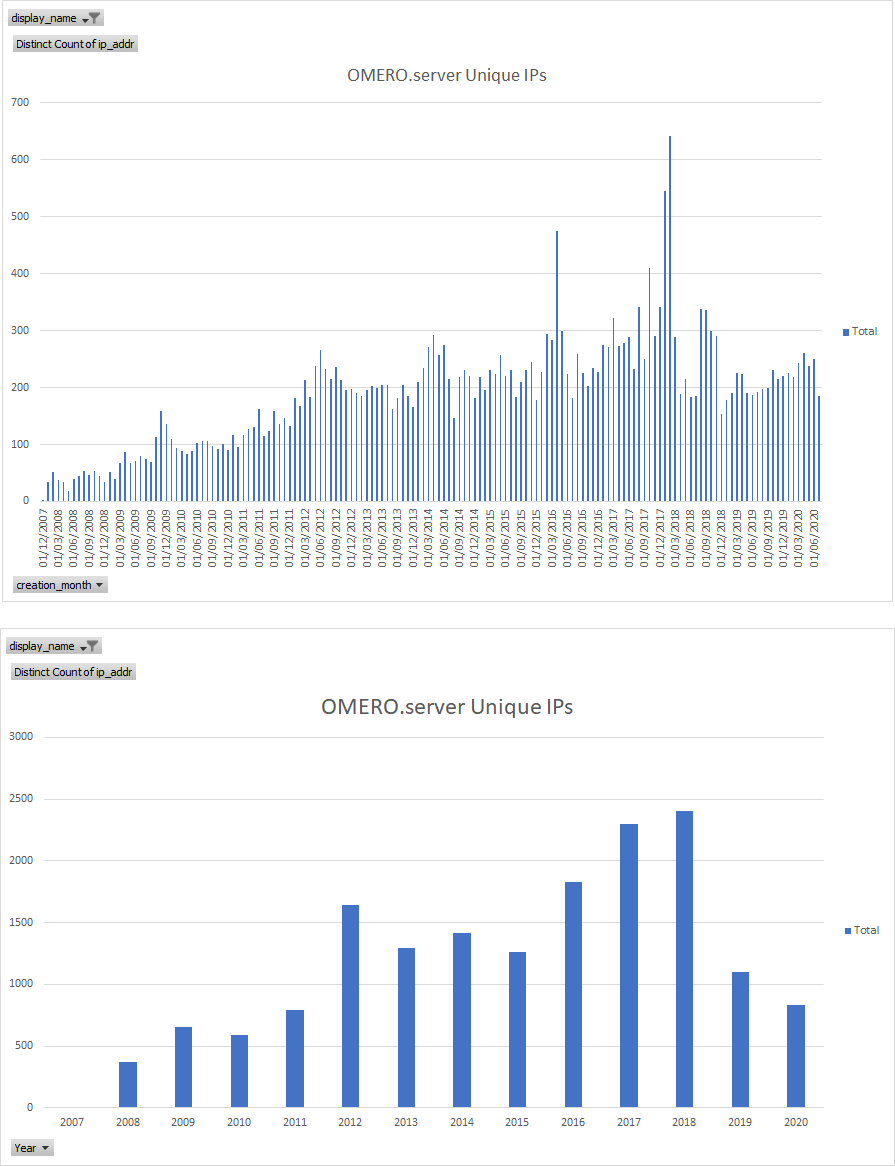
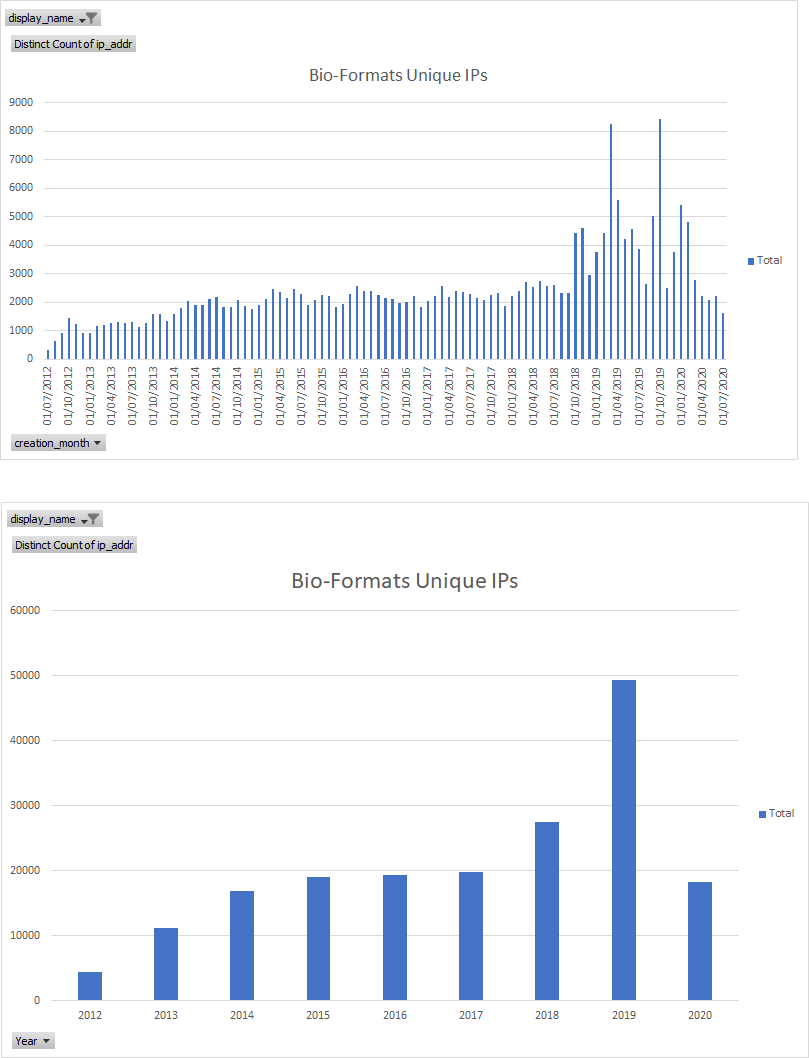
Usage Stats: upgrade checks - total vv unique IPs
Zarr fun if desired
- All PRs in ‘sets of 3’ omero-ms-zarr (spec), ome-zarr-py & omero-cli-zarr,
- ome-zarr-py #45 lots of changes. Black formatting etc. pre-commit file.
- mypi - specify return types. Work to add, but useful for refactoring
- conda environment.yml - not many versions pinned yet. Might be needed.
- testing qt via github actions is hard. OS-specific.
- ome-zarr create - to create sample data. Avoid hitting s3 in testing
- Main change: graph traversal API. entry point: parse_url()
- also split out napari logic.
- separate our metadata from napari spec. E.g. ‘contrast_limits’
- Chris: we’ve had issues with graph traversal on s3 - xarray any use?
- Josh: that’s using fsspec. Could support /add that in our api
- Layer (from napari) hard to know what’s in the tuples.
- need to add labels ‘on top’ - layer opacity allows other layers below
- Other types possible - e.g. image-image links.
- Need a name for hierarchy of data ‘layers’ with metadata
- Hierarchy is converted into ordered list of layers, retaining the relationships between them.
- Reader class - uses ‘yeild’ generators everywhere, but not used anywhere yet - napari wants everything up front.
- Scaler class - used for downsampling. Used by omero-cli-zarr so that it saves downsampled labels.
- Can now download any image with labels (both as pyramids) and open in napari.
- naming needs deciding on: e.g. labels layers are identified by ‘color’ metadata.
Expect more discussions in public when released.
Bio-Formats Total
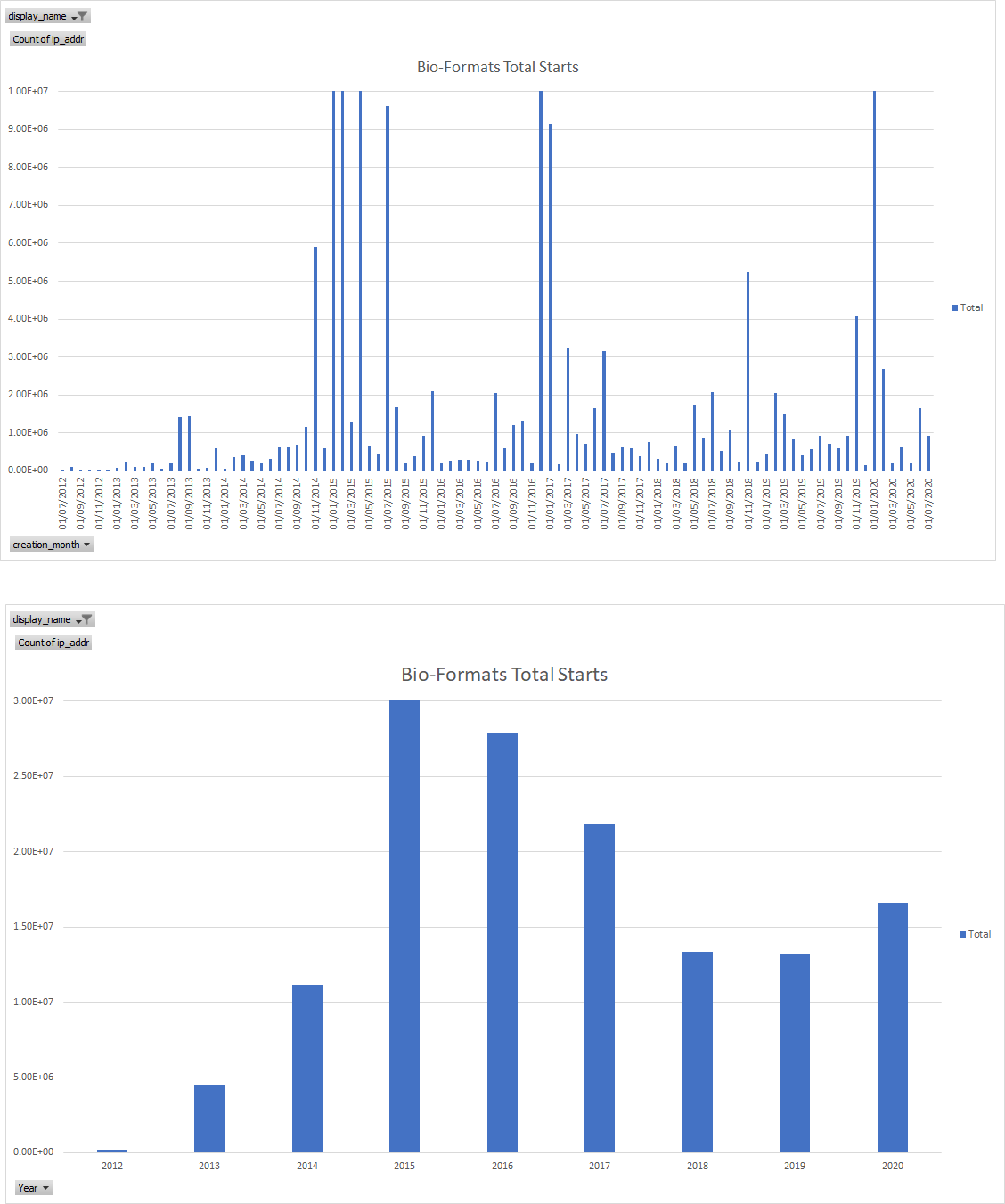
Bio-Formats Unique
OMERO Clients Total
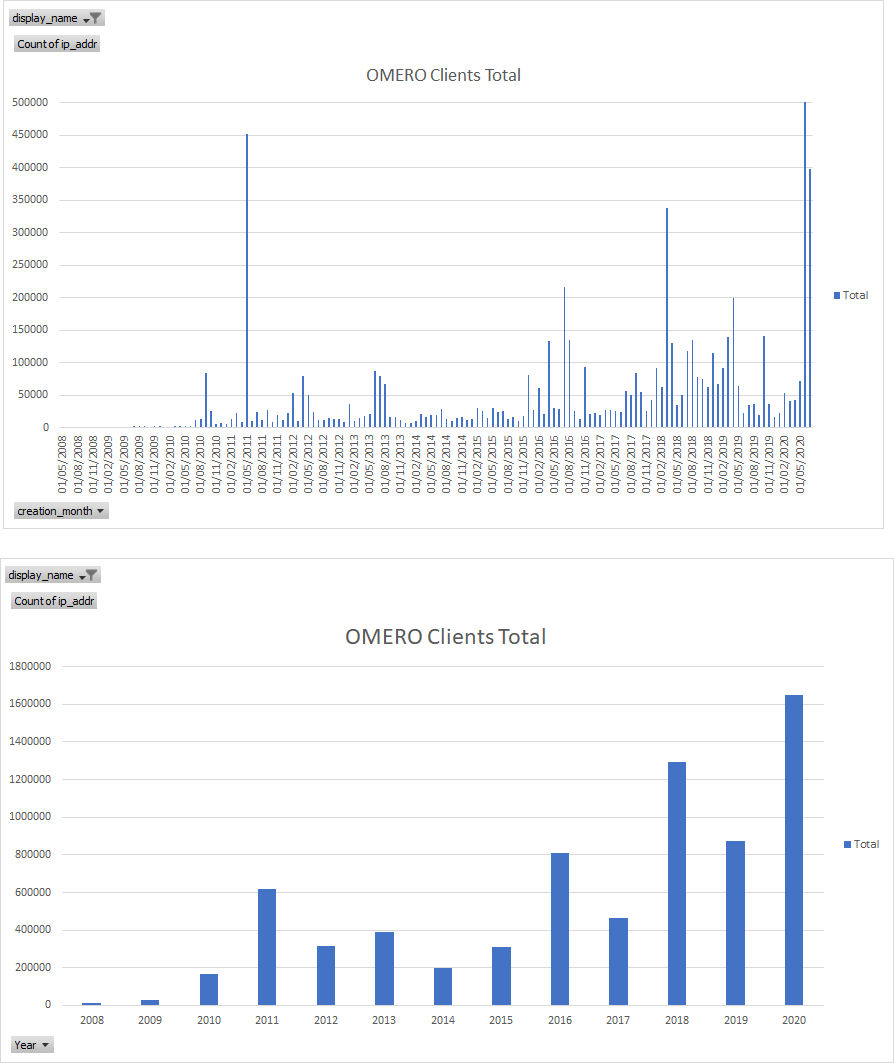
OMERO Clients Unique
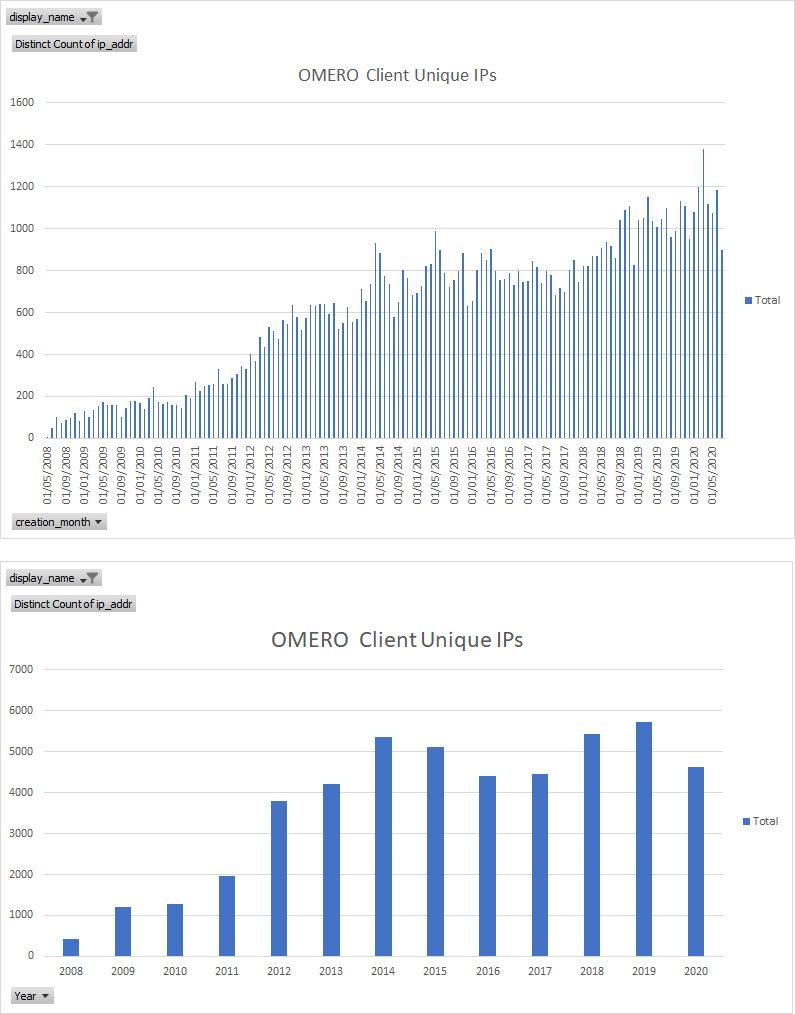
OMERO server Unique